Contents
(Source: http://epscicon.vidyaacademy.ac.in/wp-content/uploads/2017/12/eeg.jpg)
Disability diagnostic using EEG Time Series Analysis
A research approach on how to identify if a child is affected or not by autism analyzing electroencephalogram (EEG) records using Convolutional Neural Networks (CNN).
Introduction
Children can develop disabilities at birth. Identifying their problems at an early stage can lead to more consistent improvements later in life. This is mainly dependent on neuroplasticity (the brain’s ability to reorganize itself throughout life), which is much higher during the first few years of our life.
Autism Spectrum Disorder (ASD) is a type of neurodevelopmental disorder. These types of disorders are mainly considered as disruption of regular brain functioning. Over the last few years, there has been reported an increased number of children developing this pathology. Data drawn from a 2014 survey in the United States has shown an increase of 16% compared to the previous same survey conducted in 2012. According to this research, about 1 in 59 children in the United States have Autism Spectrum Disorder (“CDC’s Autism and Developmental Disabilities Monitoring (ADDM) Network”, [1]).
Recent researches have shown that intensive early start intervention approaches can lead to major improvements to this condition. Therefore, early detection using Machine Learning can play a vital role in this ambit.
In this article, I will outline different approaches I took to analyze labelled EEG data (already preprocessed) containing brainwaves records of both children affected and not by ASD. Making use of algorithms such as Support Vector Machines (SVM), Decision Trees, CNN’s and LSTM’s has been possible to achieve above 96% classification accuracy.
Machine Learning (ML)
The dataset I have been working with consisted of 129 columns. The first 128 columns represented the 128 EEG channels used during the signal acquisition, and the last column the data label (either affected or not by autism). Every 250 rows of the dataset represented a time series repetition. The time series repetitions for each child varied between 20 and 80, depending on the subject. The dataset demonstrated to be balanced, containing 49.03% of the data of children without ASD and 50.97% of the data of ASD children.
Data preprocessing consisted of first loading the data, standardizing it and then dividing it in training (70%) and test sets (30%) for both the inputs an output’s values. To do so, I made use of python libraries such as pandas, numpy, matplotlib.pyplot, sklearn and itertools. The ML models used attempted to identify if a child is affected or not by ASD using just a single stimulus repetition.
The results obtained are summarised in Table 1.
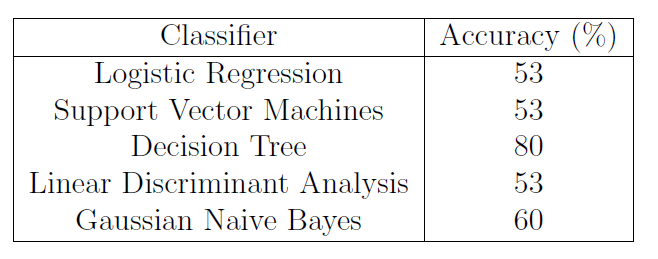
Table 1: ML Classification Accuracy.
The Decision Tree achieved the most accurate result. Decision Tree is a type of supervised learning algorithm which can be used for either classification or regression tasks. What distinguishes Decision Tree from other ML algorithms, is their ability to display their decision-making process using an upside-down tree-like representation. In the tree, each node represents a feature in the data-set, each branch a decision rule and each leaf a decision outcome. In this implementation, I decided to use the CART (Classification And Regression Tree) algorithm implementation which makes use of the Gini Index as a metric.
Convolutional Neural Networks (CNN)
In order to increase classification performances, I then decided to design a CNN model using the Keras Python library. CNN’s are a class of neural networks typically used for image recognition/classification but can be also used for time series analysis. This can be done by converting the time series in a grayscale image like format.
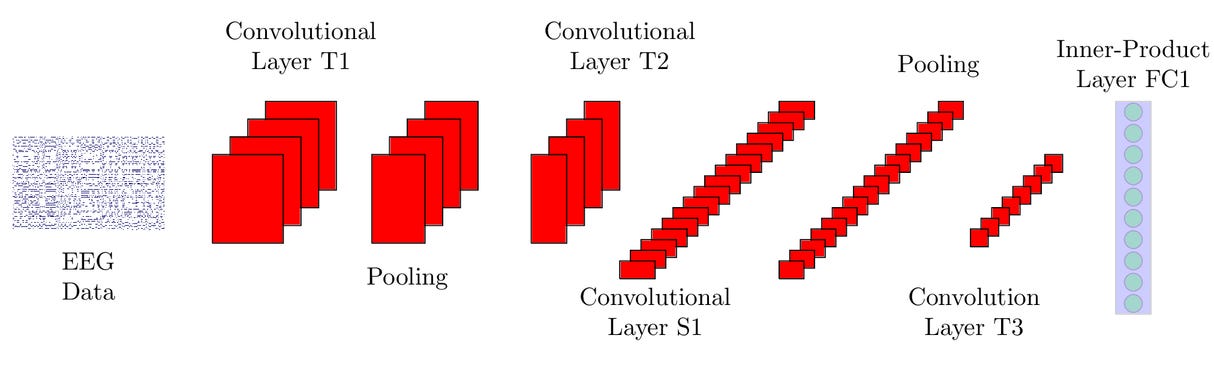 Figure 1: CNN’s in EEG analysis [2]
Figure 1: CNN’s in EEG analysis [2]
The model architecture consisted of:
- One 2D Convolutional Layer of 64 filters, a kernel size of 5x5, a ReLU (Rectified Linear Unit, Equation 1) function and same padding.
- Another 2D Convolutional Layer having 32 filters, a kernel size of 5x5, a ReLU (rectified linear unit) function, same padding and an L2 regularisation coefficient of 0.01 (to prevent overfitting).
- A 2D MaxPooling layer of 2x2 size.
- A Dropout layer of 0.2 intensity (in order to avoid overfitting the data).
- A layer to first flatten the data from three dimensions to just one, and then another one to condense the input to give to the classifier 128 features (always using the ReLU function and L2 regularisation).
- A second Dropout layer of 0.5 intensity.
- Finally, a Dense layer (of two neurons) to produce the classification result, using a Softmax activation function. The Softmax function (Equation 2) will take in this case the two inputs from the neurons and convert them in two probabilities that sum up to one. The greater probability was rounded up/down to either one or zero to represent the CNN output choice.
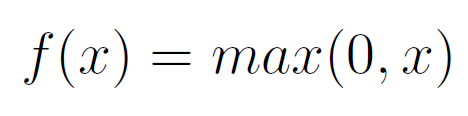
Equation 1: ReLU Activation Function

Equation 2: Softmax Function
In order to optimize the training, the Adam (Adaptive Moment Estimation) gradient descent algorithm was used, and the cross-entropy function was used to compute the model loss. The cross-entropy function, in a binary classification case, can be calculated by using Equation 3.
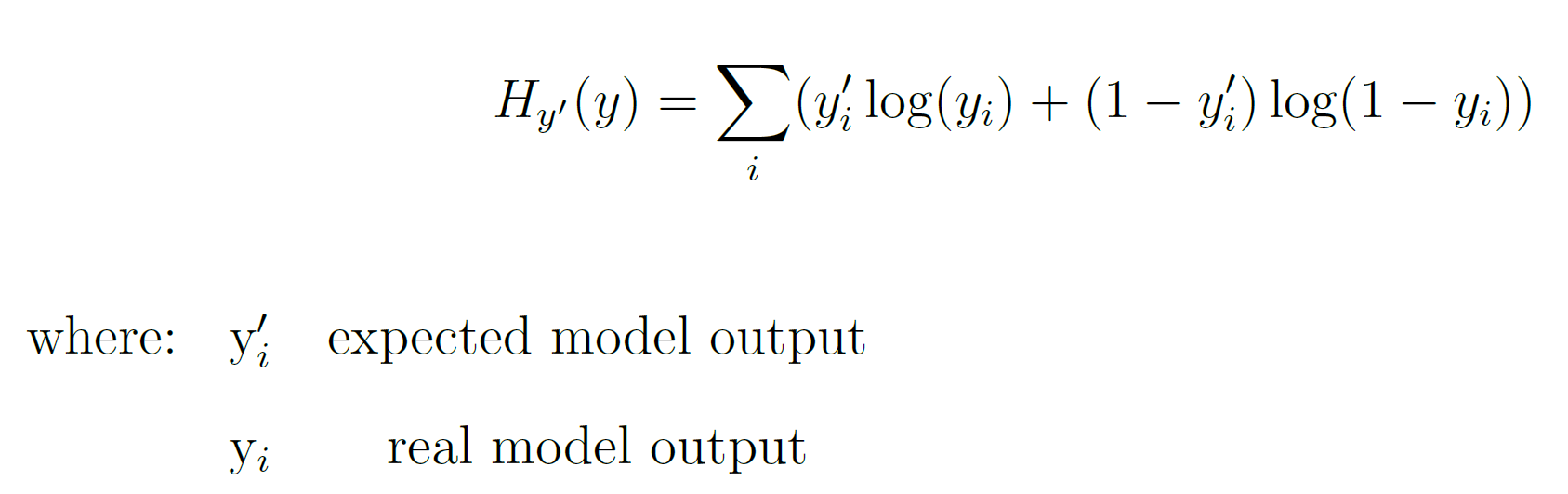
Equation 3: Cross-Entropy Function
This model achieved an overall validation accuracy of 94.58% in just thirty epochs of training.
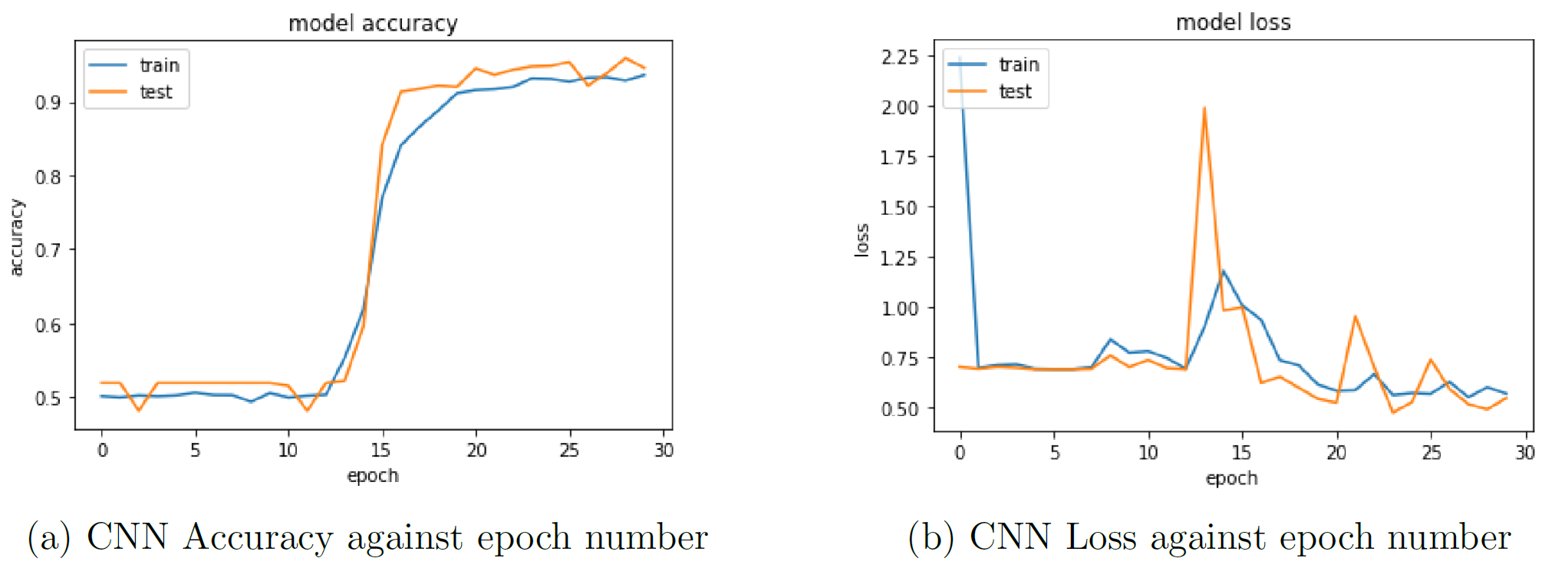
Figure 2: CNN accuracy and loss
Conclusion
This article demonstrated that is overall possible to achieve good classification results using ML and Deep Learning models on preprocessed EEG data. As a further development of this project, I also managed to develop an LSTM model able to achieve over 96% classification accuracy. Other metrics such as Confusion Matrix, Sensitivity, Specificity, and AUC-ROC Curve have been used to cross-validate the accuracy of these models.
As an addition, would be interesting to apply an Auto-Encoder Network [3] during the pre-processing stage to examine if that could make classification easier and speed up the model training time.
One issue to be taken into account in this experiment could be the reliability of the data used (eg. if the data was collected correctly, if all the noise registered during the measurements had been correctly filtered out…) and how reproducible these accuracy results can be using other forms of data which have been collected using different types of EEG Cap or under different laboratory conditions.
When trying to use Artificial Intelligence in medical applications, high accuracy and reliability must be obtained. Explainable AI (creating models able to explain himself their classification decision making process) could help doctors to understand how/if to use AI when making decisions.
I would like to thank Professor Koushik Maharatna for giving me the opportunity to carry out this project.
Bibliography
[1] Division of Birth Defects, National Center on Birth Defects and Developmental Disabilities, Centers for Disease Control and Prevention. Accessed: https://www.cdc.gov/ncbddd/autism/data.html December 2018.
[2] Single-trial EEG RSVP classification using convolutional neural networks, Jared Shamwell, Hyungtae Lee et al. Accessed: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/9836/983622/Single-trial-EEG-RSVP-classification-using-convolutional-neural-networks/10.1117/12.2224172.short?SSO=1 June 2019.
[3] Jeremy Jordan Introduction to autoencoders. Accessed: https://www.jeremyjordan.me/autoencoders/ March 2019.
Contacts
If you want to keep updated with my latest articles and projects follow me on Medium and subscribe to my mailing list. These are some of my contacts details:
